Open Source Tools and Resources
We strongly believe in global access to open and reproducible science. Our tools, software, and resources are openly licensed and freely available for all to use and build upon.
Featured
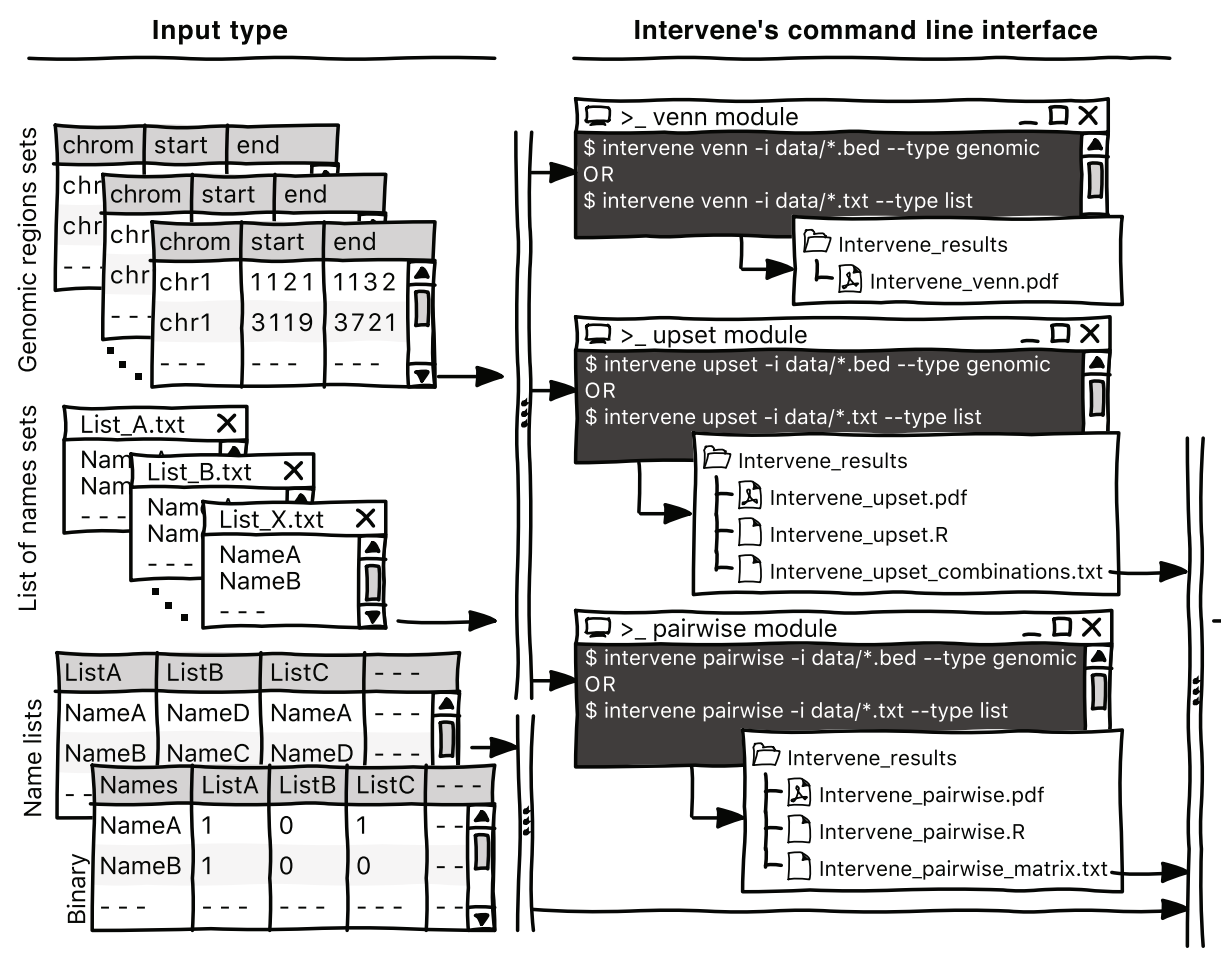
Intervene
A tool for intersection and visualization of multiple gene or genomic region sets

JASPAR database
The open and high-quality transcription factor binding profile database
More

UniBind
A map of direct TF-DNA interactions in the human genome.
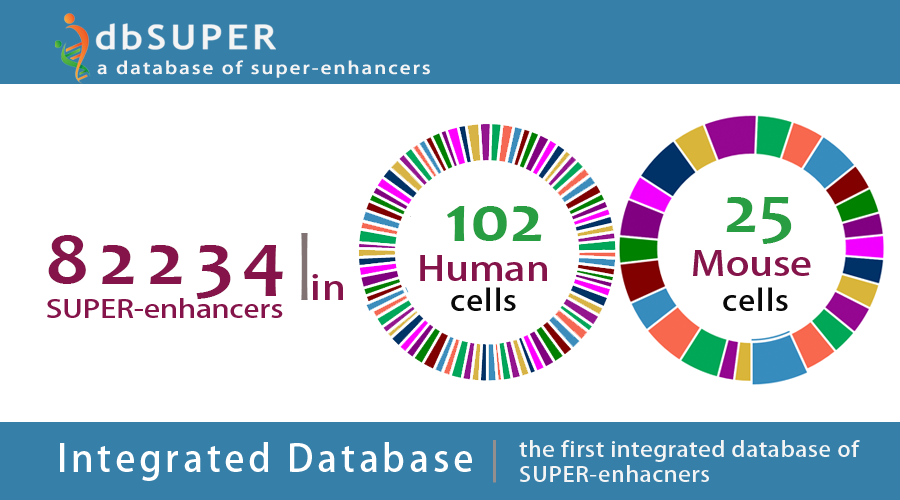
dbSUPER
The first integrated and interactive database of super-enhancers

JASPAR RESTful API
Programmatically accessing JASPAR data from any programming language

mCrossBase
A database of RNA-binding proteins binding motifs and crosslink sites

ith.Variant
somatic variant detection from multi-sampled genomic WGS/WES data of tumors

BiasAway
A tool to generate composition-matched background sequence set

ECRcentral
A central platform for early career researchers community

imPROSE
an integrated methods to predict super-enhancers

pyJASPAR
A Pythonic interface to JASPAR transcription factor motifs
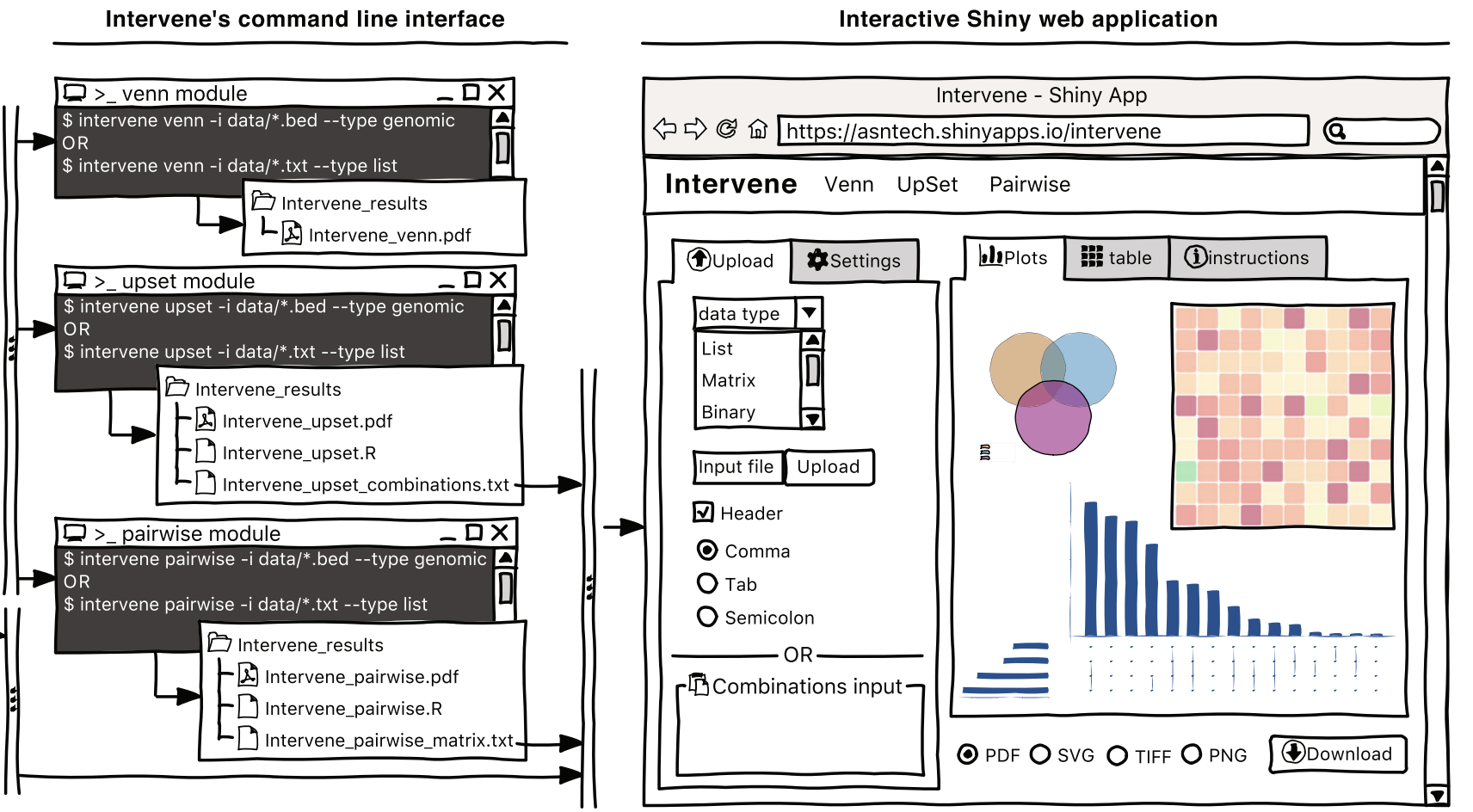
Intervene ShinyApp
A tool for intersection and visualization of multiple gene or genomic region sets
